The small-but-perfectly-formed Genome technical team have spent the last six months digging into the vast cache of data that was produced during the scanning over 350,000 pages of the Radio Times. They’ve found quite a lot interesting artefacts in unexpected places, and are currently brushing them down and cleaning them up in readiness for publication on the site...
I’ll describe a couple of these artefacts, and try to explain how they came to exist and what we are doing with them.
The Genome project set out to create an easily navigable and searchable broadcast history of the �������� - the public face of the ��������’s catalogue - rather than to simply digitise the Radio Times. This meant that substantial chunks of the magazines would not be made available online (e.g the adverts, articles, letters), as these would have made the listings themselves harder to find.
In order to achieve this, during the design of the data extraction process, algorithms were written to automagically identify which sections of the magazine and which elements within a page were likely to contain a listing (as opposed to anything else). These algorithms essentially looked for patterns in the layout of pages and patterns in sequences of characters.
As with most automated pattern recognition scenarios, there will always be an element of error. Given the substantial variance in the design and layout of the Radio Times between 1923 and 2009 (I blame the graphic designers) these algorithms had to be continually tweaked to achieve a satisfactory accuracy rate, but a degree of error was inevitable. Finding and correcting these errors is of huge importance if we are going to be able to make available the fullest and most accurate representation of the ��������’s broadcast history.
Simon Smith - our champion data archaeologist - spent a substantial amount of time poring over a million or so rows of data in the Genome database in two tables, called ‘genome_related’ and' genome_tables’. Fields in these rows contain blocks of text that the algorithms decided were not programme listings (it usually decided that they were articles and therefore not for publication).
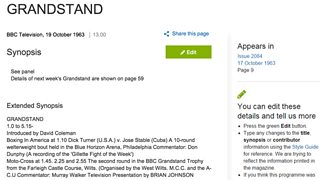
This is what a Grandstand listing would look like.
Simon found that in many cases, these fields contained either what was essentially an extended synopsis (as found in text boxes adjacent to the programme listing or in the longer photo captions) or further details about the programme listed (such as which pieces by which composer would be played during a radio music programme) or ‘sub-listings’ (such as the start time of the various sports featured in an episode of Grandstand). He has painstakingly identified more than a hundred thousand items which we are working on making available on the Genome website, adding a whole new level of detail to the listings.
We are very excited (yes - we do need to get out more) about something else that Simon has found: around 150,000 chunks of text that the algorithms misidentified as articles when in fact they are brief programme listings. We estimate that when the data is extracted and properly classified, we might be able to add nearly a million new programme listings to Genome. This will take a good while to get ready, but we’ll blog about it in more detail when we have made some more progress.
